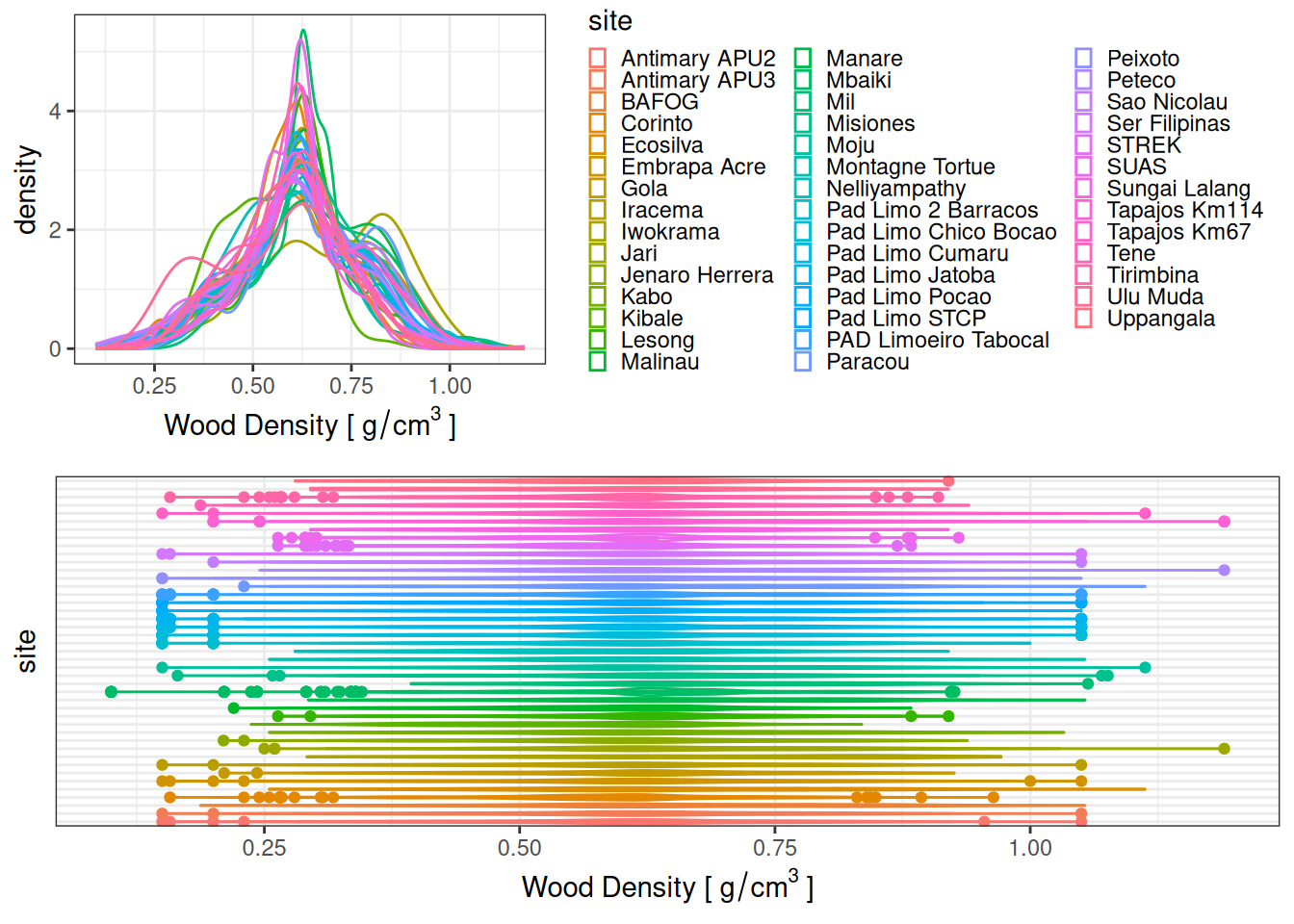
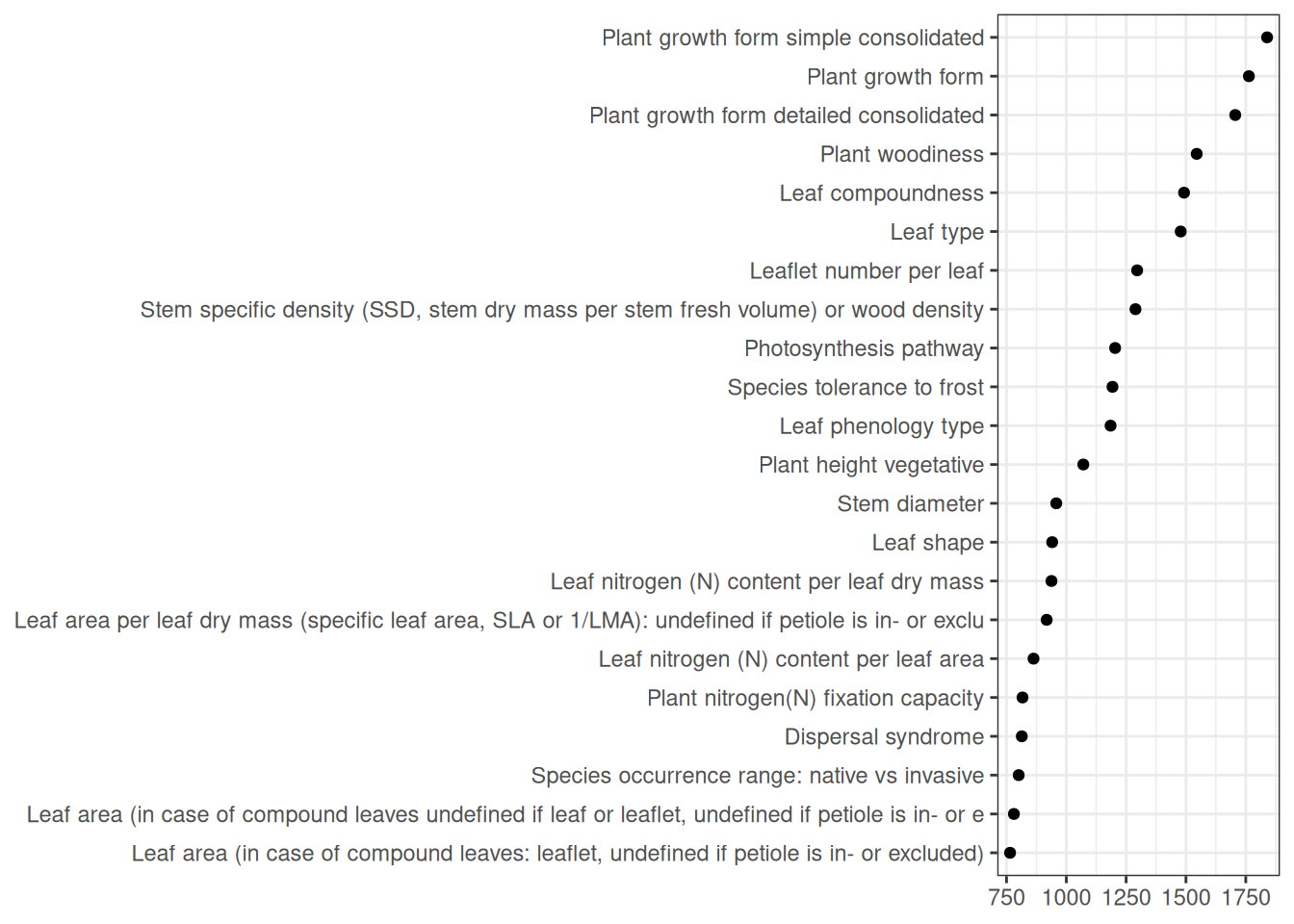
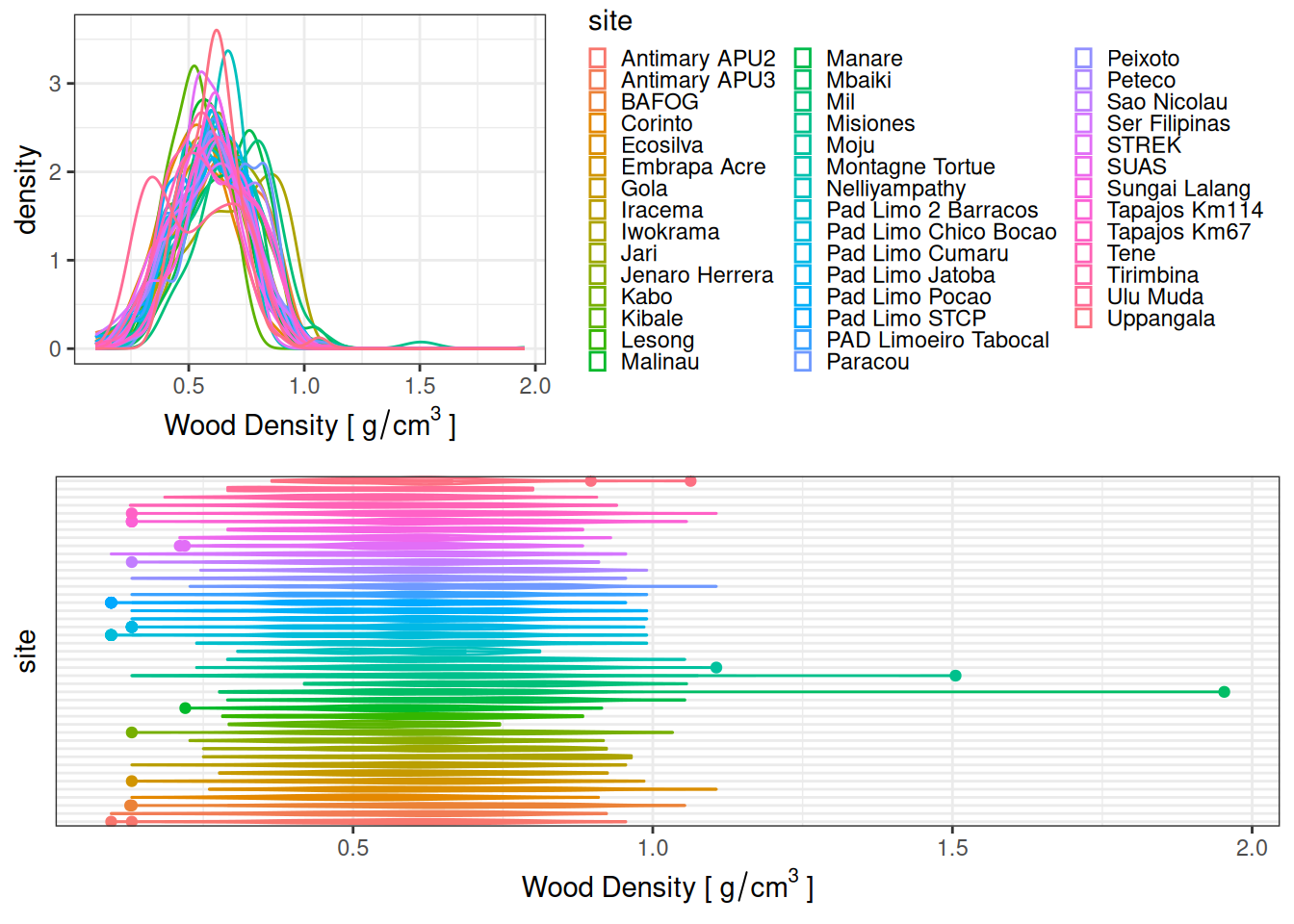
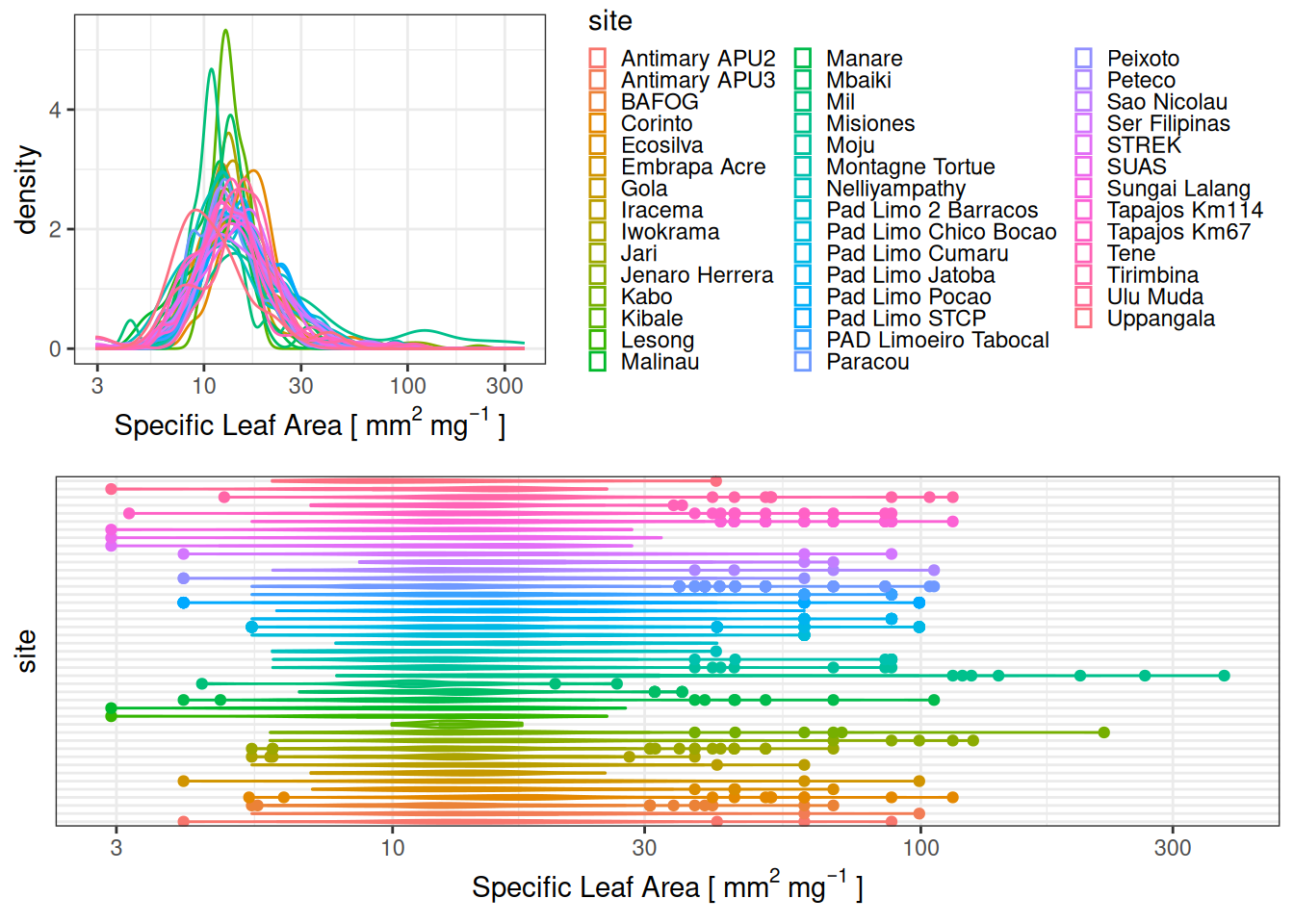
| 1 |
53035, 8582, 14866, 53654, 14156, 15557, 27269, 56630, 29126, 52505, 31260, 30552, 4136, 17195, 8580, 32629, 39236, 8980, 3147, 86888, 52969, 38544, 30609, 27124, 36681, 39724, 26812, 40311, 36684, 37540, 46572, 34387, 44223, 22256, 49908, 52432, 13692, 430788, 33337, 44232, 5583, 30895, 12681, 43766, 54284, 30632, 33380, 52899, 246054, 56788, 12678, 43849, 11219, 14450, 18719, 14882, 56617, 33288, 24884, 38616, 34870, 20736, 408207, 50321, 54536, 54783, 103114, 40645, 35216, 30904, 44986, 25648, 52844, 40368, 41051, 22281, 32634, 430797, 30624, 52895, 44853, 37333, 43959, 43542, 33334, 21888, 26519, 38552, 45280, 29808, 20479, 15619, 40831, 19480, 55518, 47684, 19696, 55524, 36897, 44855, 55866, 56813, 52968, 92496, 43763, 4160, 7610, 10273, 17778, 40357, 11353, 56697, 27177, 57266, 38516, 53047, 29162, 22272, 50499, 46274, 54663, 43873, 57627, 57282, 52436, 30575, 14860, 43767, 39875, 32613, 14890, 21873, 99866, 29834, 74946, 5605, 52900, 12272, 33343, 27264, 8569, 39234, 42597, 84130, 10716, 40823, 40360, 11365, 40231, 11345, 44590, 43958, 37888, 8576, 18800, 101052, 53039, 35441, 32627, 3383, 50390, 39604, 29655, 57424, 56781, 35358, 55852, 35649, 54098, 11952, 3878, 34856, 51769, 36686, 38676, 38574, 3877, 50523, 33335, 5937, 6614, 19578, 22258, 52390, 3446, 51785, 22261, 38551, 43836, 33295, 43828, 13282, 32615, 50507, 54280, 29811, 43810, 19522, 30614, 2458 |