Code
library(tidyverse)
library(knitr)library(tidyverse)
library(knitr)Below we show the productivity in four sites that have been remeasured for multiple decades.
data <- read.csv("data/derived_data/aggregated_data_v9.csv") |>
mutate(plot_new = Plot) |>
separate(Plot, "Plot", "_")
plot_info <- read.csv("data/raw_data/bioforest-plot-information.csv")data |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
subset(variable %in% c("agb_growth", "agb_recr")) |>
subset(Site %in% c("Mbaiki", "Paracou", "Ulu Muda", "Lesong")) |>
pivot_wider(names_from = "variable") |>
ggplot(aes(Year, agb_recr + agb_growth, ,
group = plot_new,
col = Treatment
)) +
geom_point() +
geom_line() +
labs(y = "AGB productivity [Mg/ha/yr]", col = NULL) +
facet_wrap(~Site, scales = "free") +
theme_minimal()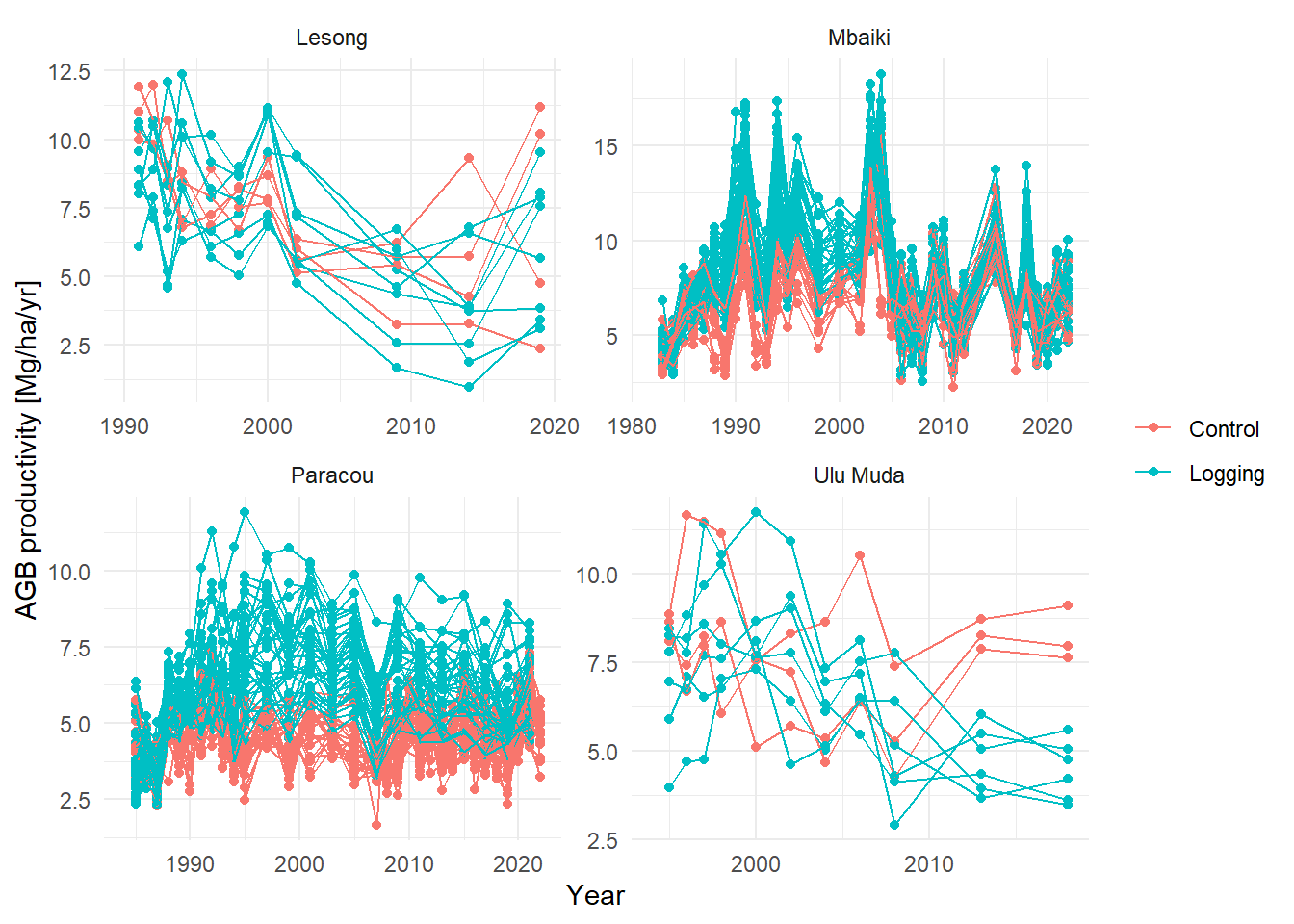
data |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
subset(variable == "nstem_recr") |>
subset(Site %in% c("Mbaiki", "Paracou", "Ulu Muda", "Lesong")) |>
ggplot(aes(Year, value, group = plot_new, col = Treatment)) +
geom_point() +
geom_line() +
labs(y = "Stem recruitment [/ha/yr]", col = NULL) +
facet_wrap(~Site, scales = "free") +
theme_minimal()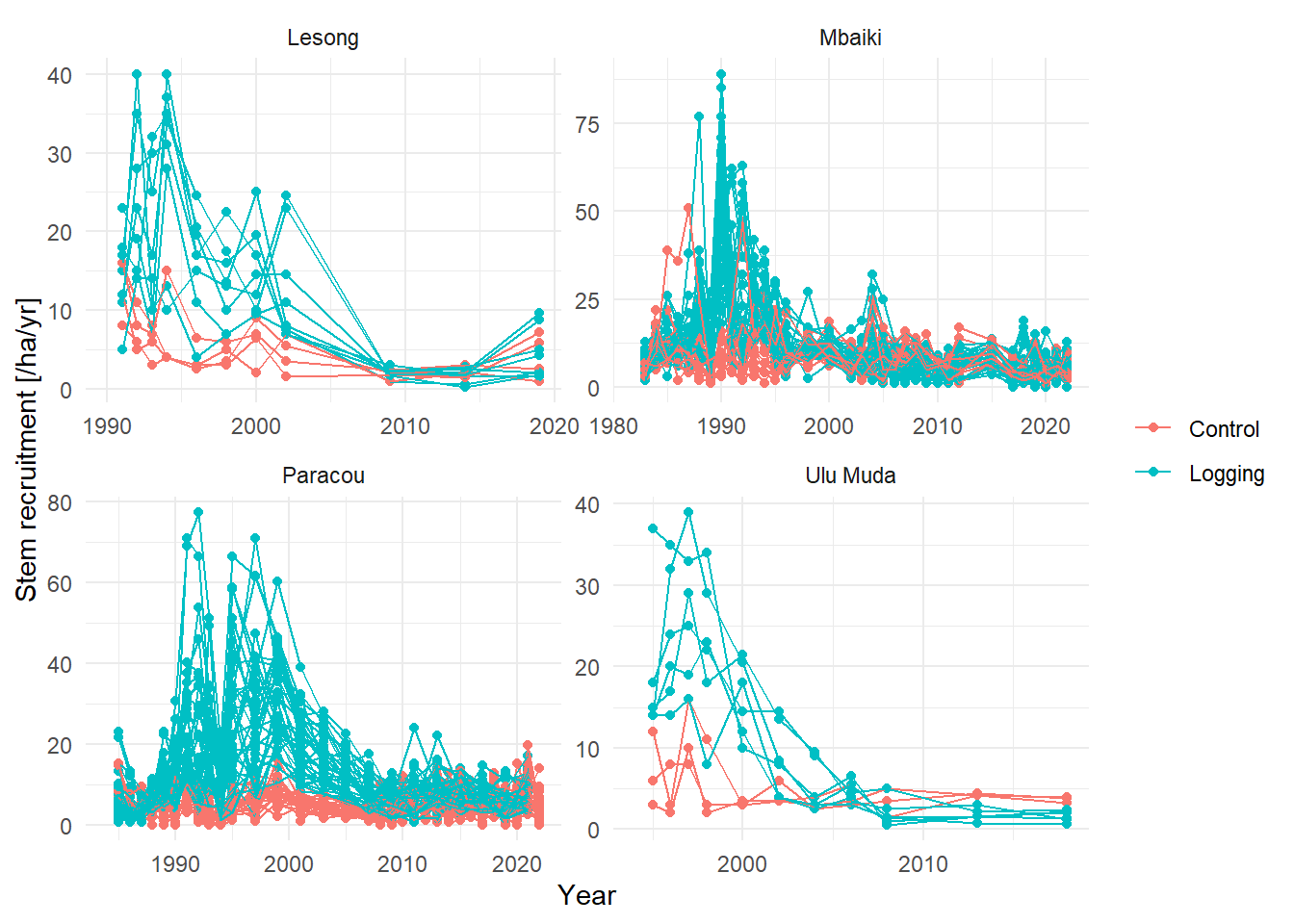
data |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
subset(variable == "agb_mort") |>
filter(Treatment == "Control" | Year >= Year_of_harvest + 3) |>
subset(Site %in% c("Mbaiki", "Paracou", "Ulu Muda", "Lesong")) |>
ggplot(aes(Year, value, group = plot_new, col = Treatment)) +
geom_point() +
geom_line() +
labs(y = "AGB mortality [Mg/ha/yr]", col = NULL) +
facet_wrap(~Site, scales = "free") +
theme_minimal()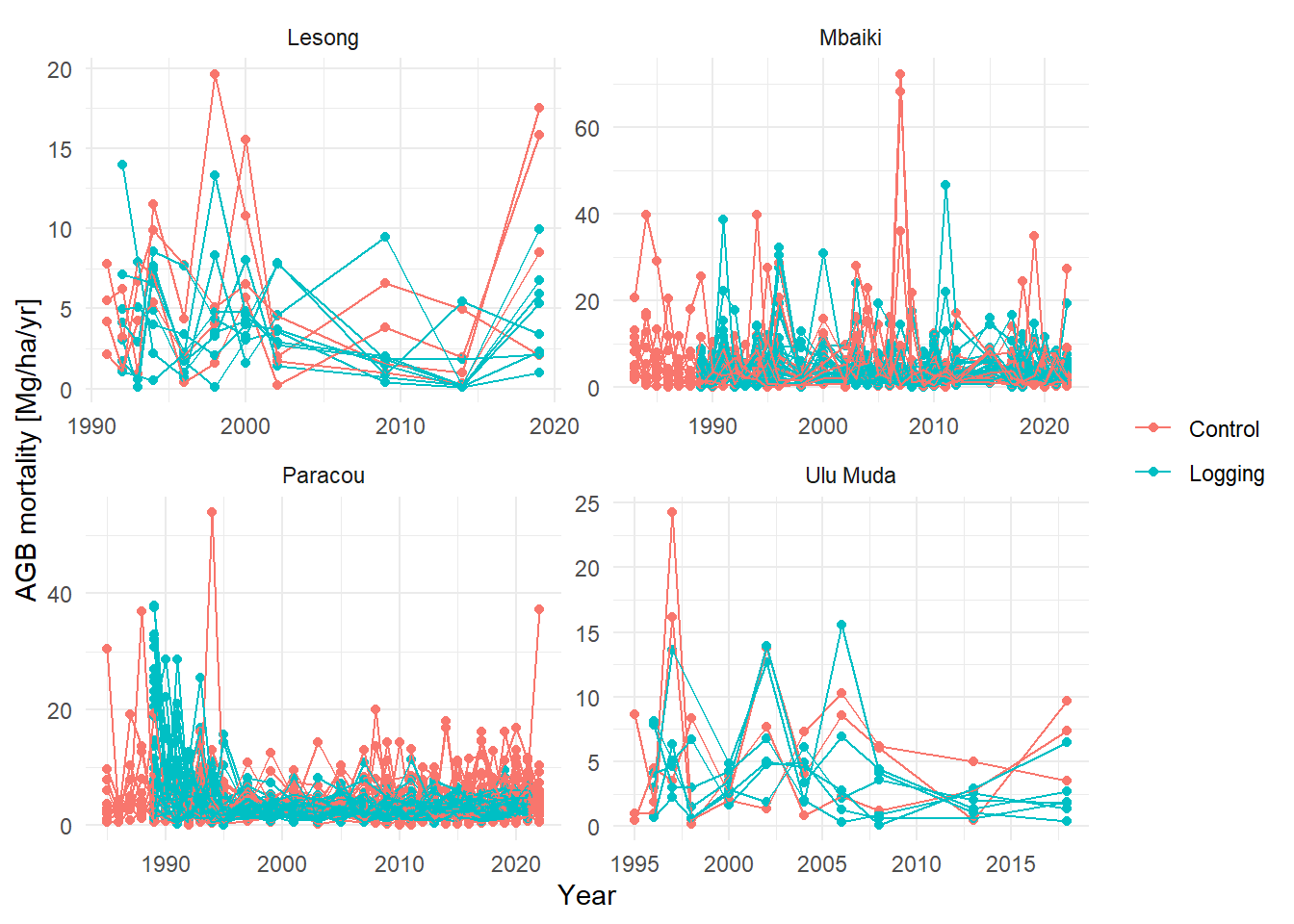
data |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
subset(variable == "nstem_mort") |>
filter(Treatment == "Control" | Year >= Year_of_harvest + 3) |>
subset(Site %in% c("Mbaiki", "Paracou", "Ulu Muda", "Lesong")) |>
ggplot(aes(Year, value, group = plot_new, col = Treatment)) +
geom_point() +
geom_line() +
labs(y = "Stem mortality [/ha/yr]", col = NULL) +
facet_wrap(~Site, scales = "free") +
theme_minimal()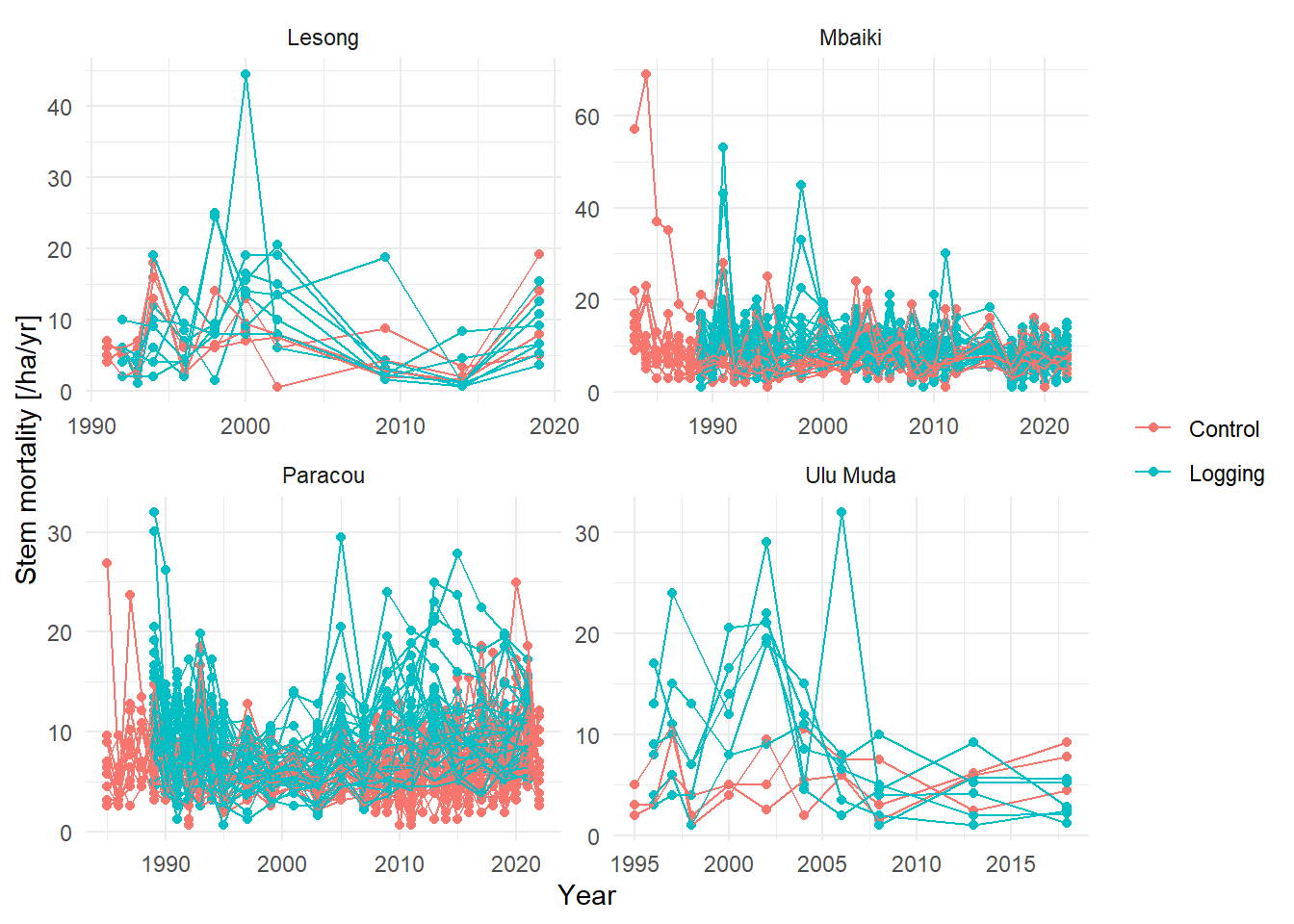
In this analysis, we only kept sites that meet the following criteria:
at least 4 post-logging measurements
at least 10 years of post-logging measurements
presence of control plots
productivity values < 10% (too high)?
criteria_site <- data |>
inner_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
group_by(Site) |>
summarise(
controls = length(unique(plot_new[Treatment == "Control"])),
logged = length(unique(plot_new[Treatment == "Logging"]))
)
data |>
inner_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
filter(Treatment == "Logging" & !is.na(Year)) |>
group_by(Site, plot_new) |>
summarise(
ncensus = length(unique(Year[Year > Year_of_harvest])),
tcensus = max(Year, na.rm = TRUE) - unique(Year_of_harvest)
) |>
group_by(Site) |>
summarise(ncensus = median(ncensus), tcensus = median(tcensus)) |>
left_join(criteria_site) |>
filter(ncensus > 3, tcensus > 9, controls > 0, logged > 0) |>
write.csv("data/derived_data/sites_to_keep.csv", row.names = FALSE)read.csv("data/derived_data/sites_to_keep.csv") |>
arrange(-ncensus) |>
kable()| Site | ncensus | tcensus | controls | logged |
|---|---|---|---|---|
| Mbaiki | 30 | 36.0 | 12 | 28 |
| Paracou | 22 | 35.0 | 24 | 36 |
| Lesong | 12 | 30.0 | 4 | 8 |
| SUAS | 12 | 24.0 | 4 | 16 |
| Ulu Muda | 12 | 25.0 | 3 | 6 |
| Sungai Lalang | 11 | 27.0 | 3 | 6 |
| Tapajos Km67 | 11 | 36.0 | 18 | 48 |
| Corinto | 10 | 29.0 | 3 | 6 |
| BAFOG | 9 | 74.0 | 8 | 12 |
| Jenaro Herrera | 8 | 28.0 | 2 | 7 |
| Jari | 7 | 26.0 | 4 | 36 |
| Misiones | 7 | 19.5 | 4 | 14 |
| Kibale | 6 | 52.0 | 11 | 15 |
| STREK | 6 | 33.0 | 12 | 36 |
| Kabo | 4 | 33.0 | 3 | 9 |