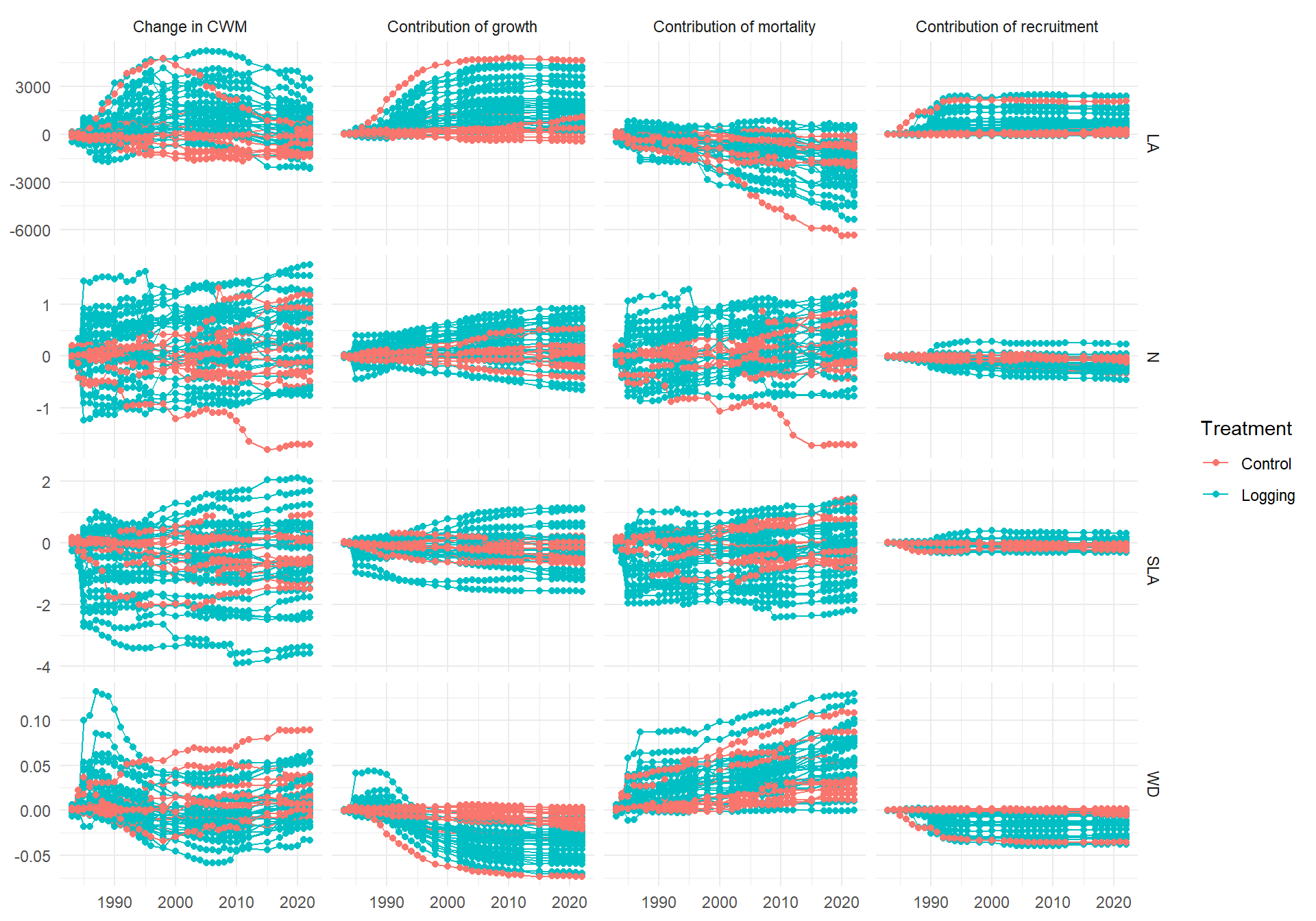
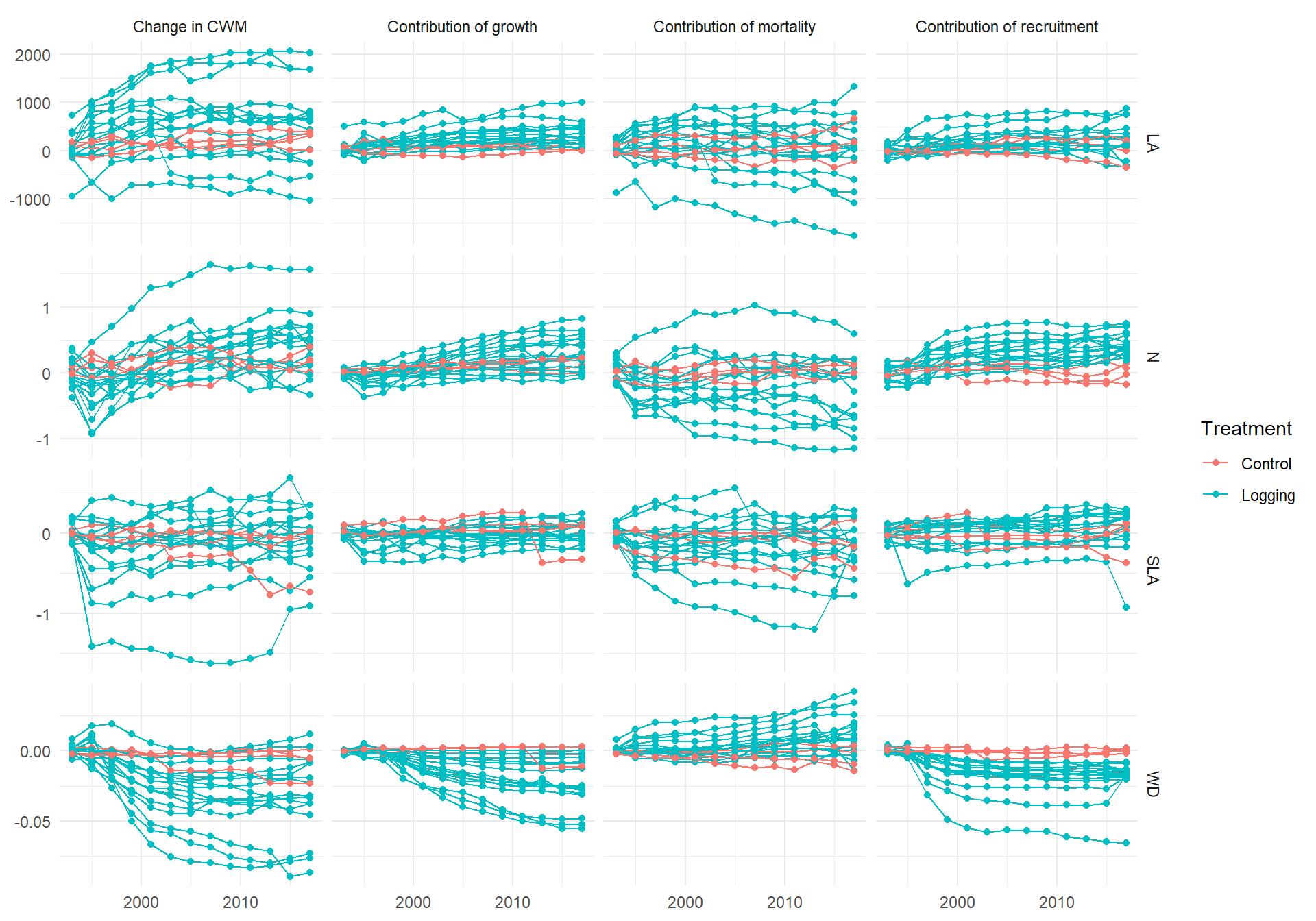
Code
plot_info <- read.csv("data/raw_data/bioforest-plot-information.csv") |>
select(site, plot, Year_of_harvest, Treatment)
data <- read.csv("data/derived_data/aggregated_data_v9.csv") |>
mutate(plot_new = Plot) |>
separate(Plot, "Plot", "_") |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
filter(grepl("cwmdiff", variable)) |>
separate(variable, c("trait", NA, "flux"), sep = "_")