We retrieved all data from SoilGrids detailed below (Poggio et al. 2021 ) :
Clay: Proportion of clay particles (< 0.002 mm) in the fine earth fraction, %
Sand: Proportion of sand particles (> 0.05 mm) in the fine earth fraction, %
Silt: Proportion of silt particles (≥ 0.002 mm and ≤ 0.05 mm) in the fine earth fraction, %
BD: Bulk density of the fine earth fraction, kg dm³
CEC: Cation Exchange Capacity of the soil, cmolC kg-1
CF: Volumetric fraction of coarse fragments (> 2 mm), cm3 100cm-3
N: Total nitrogen, g kg-1
pH: Soil pH
SOC: Soil organic carbon content in the fine earth fraction, g kg-1
OCD: Organic carbon density, kg dm³
OCS: Organic carbon stocks, kg m2
Code
list.files ("data/derived_data/soil/" , full.names = TRUE ) %>% read_tsv () %>% select (- time) %>% gather (variable, value, - site, - X, - Y) %>% separate (variable, c ("variable" , "depth" ), "_" ) %>% separate (depth, c ("mindepth" , "depth" ), "-" ) %>% mutate (depth = as.numeric (gsub ("cm" , "" , depth))) %>% select (- mindepth) %>% pivot_wider (names_from = variable, values_from = value) %>% mutate (bdod = bdod / 100 ,cec = cec / 10 ,cfvo = cfvo / 10 ,clay = clay / 10 ,sand = sand / 10 ,silt = silt / 10 ,nitrogen = nitrogen / 100 ,phh2o = phh2o / 10 ,soc = soc / 10 ,ocd = ocd / 10 ,ocs = ocs / 10 %>% write_tsv ("data/derived_data/soil.tsv" )
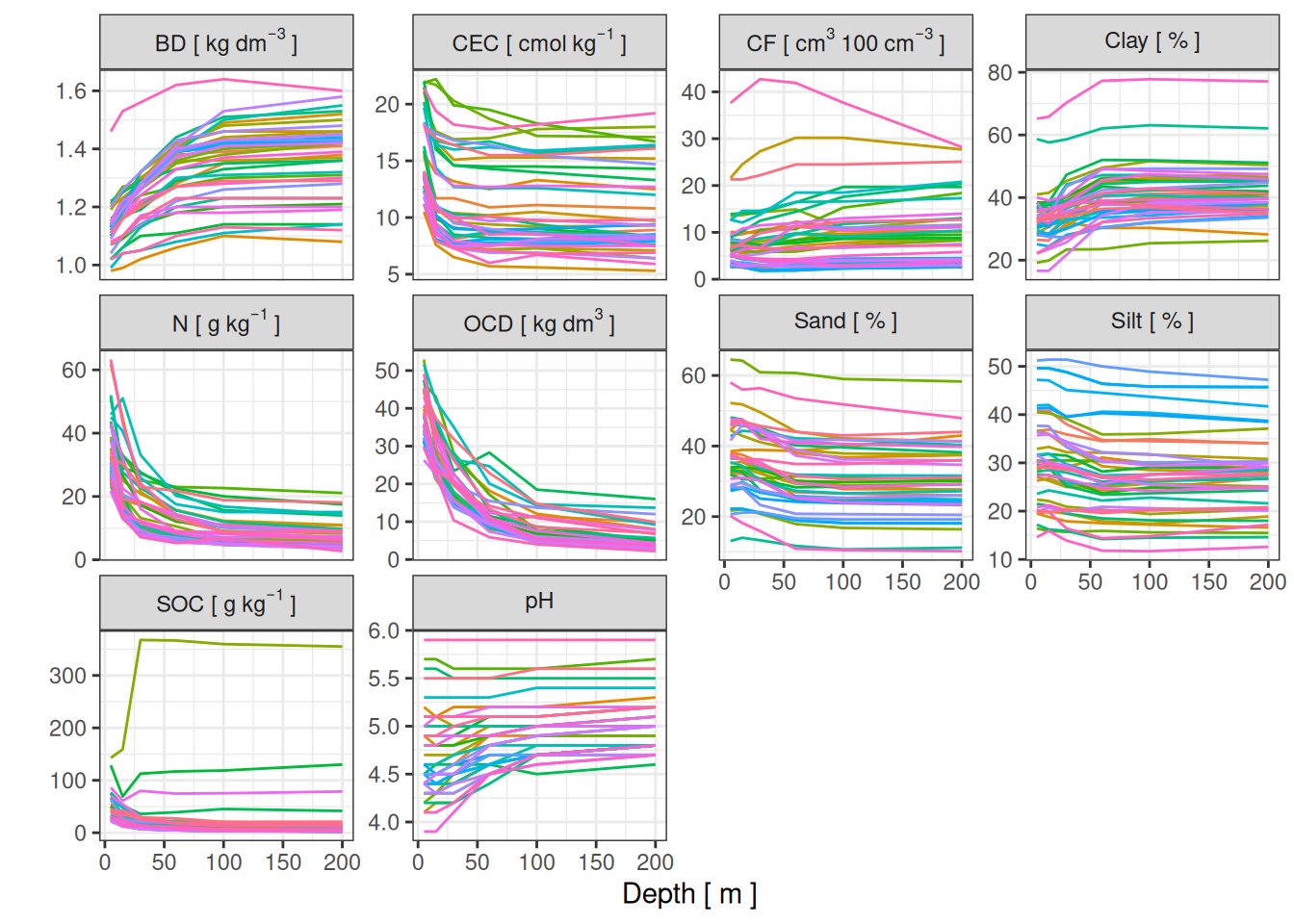
We have information per depth in each plot as shown below, I focused on the two top horizons (0-15cm) revealing most inter-site variations (V. Freycon pers. com.).
Code
read_tsv ("data/derived_data/soil.tsv" ) %>% select (- X, - Y, - ocs) %>% gather (variable, value, - depth, - site) %>% mutate (var_long = recode (variable,"bdod" = '"BD ["~kg~dm^{-3}~"]"' ,"cec" = '"CEC ["~cmol~kg^{-1}~"]"' ,"cfvo" = '"CF ["~cm^3~"100"~cm^{-3}~"]"' ,"clay" = '"Clay ["~"%"~"]"' ,"nitrogen" = '"N ["~g~kg^{-1}~"]"' ,"ocd" = '"OCD ["~kg~dm^{3}~"]"' ,"phh2o" = "pH" ,"sand" = '"Sand ["~"%"~"]"' ,"silt" = '"Silt ["~"%"~"]"' ,"soc" = '"SOC ["~g~kg^{-1}~"]"' %>% ggplot (aes (depth, value, colour = site)) + geom_line () + facet_wrap (~ var_long, scales = "free_y" , labeller = label_parsed) + theme_bw () + ylab ("" ) + xlab ("Depth [ m ]" ) + scale_color_discrete (guide = "none" )
The plots showed low variation within site but wider variation among sites.
Code
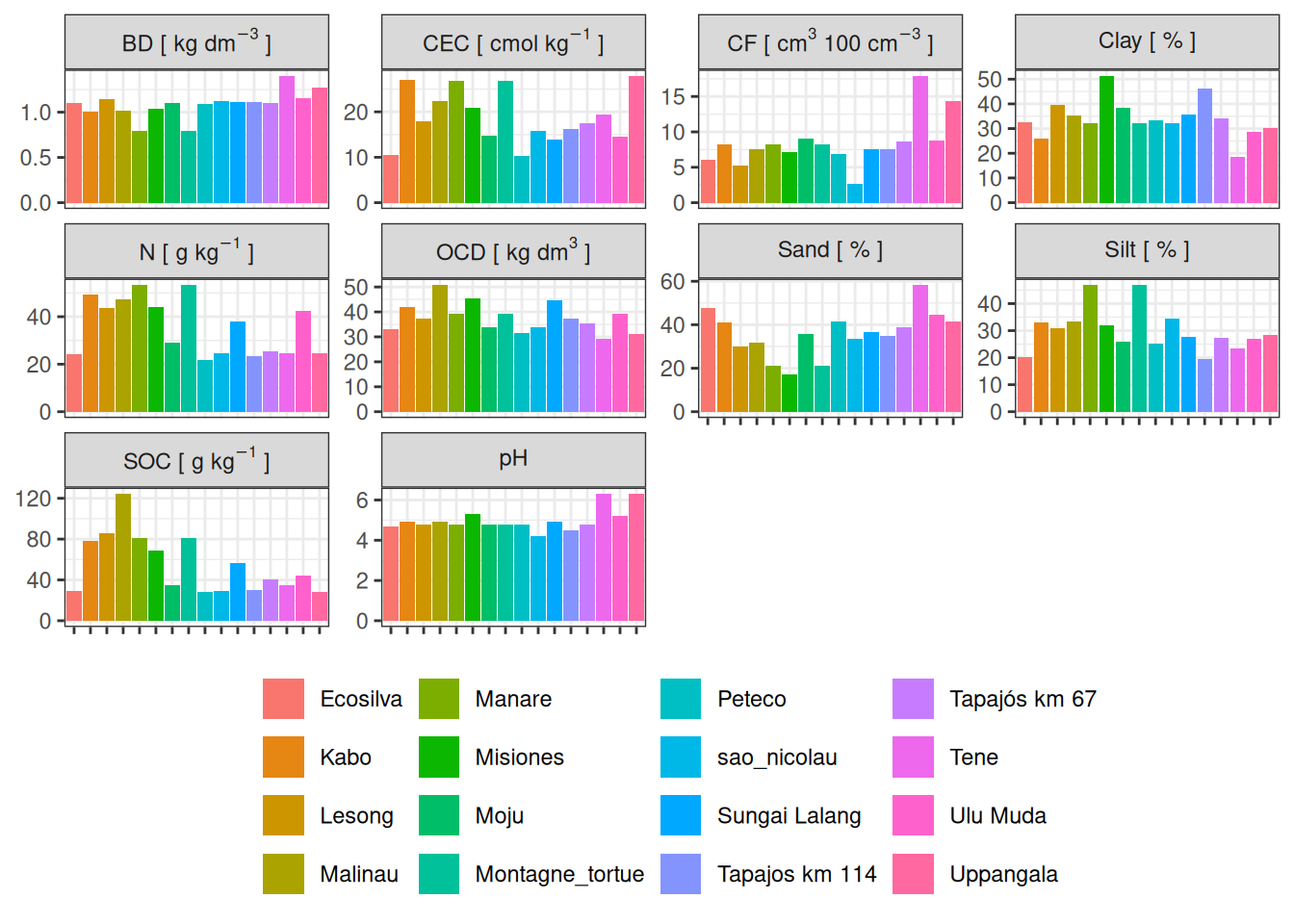
read_tsv ("data/derived_data/soil.tsv" ) %>% select (- X, - Y) %>% group_by (site) %>% filter (depth <= 15 ) %>% summarise_all (mean, na.omit = TRUE ) %>% gather (variable, value, - site, - depth) %>% na.omit () %>% mutate (var_long = recode (variable,"bdod" = '"BD ["~kg~dm^{-3}~"]"' ,"cec" = '"CEC ["~cmol~kg^{-1}~"]"' ,"cfvo" = '"CF ["~cm^3~"100"~cm^{-3}~"]"' ,"clay" = '"Clay ["~"%"~"]"' ,"nitrogen" = '"N ["~g~kg^{-1}~"]"' ,"ocd" = '"OCD ["~kg~dm^{3}~"]"' ,"ocs" = '"OCS ["~kg~m^{-2}~"]"' ,"phh2o" = "pH" ,"sand" = '"Sand ["~"%"~"]"' ,"silt" = '"Silt ["~"%"~"]"' ,"soc" = '"SOC ["~g~kg^{-1}~"]"' %>% ggplot (aes (site, value, fill = site)) + geom_col (aes (group = as.factor (depth)), position = "dodge" ) + facet_wrap (~ var_long, scales = "free_y" , labeller = label_parsed) + theme_bw () + theme (legend.position = "bottom" , axis.title = element_blank (),axis.text.x = element_blank (),legend.key.size = unit (0.5 , "line" )+ scale_fill_discrete ("" )
Code
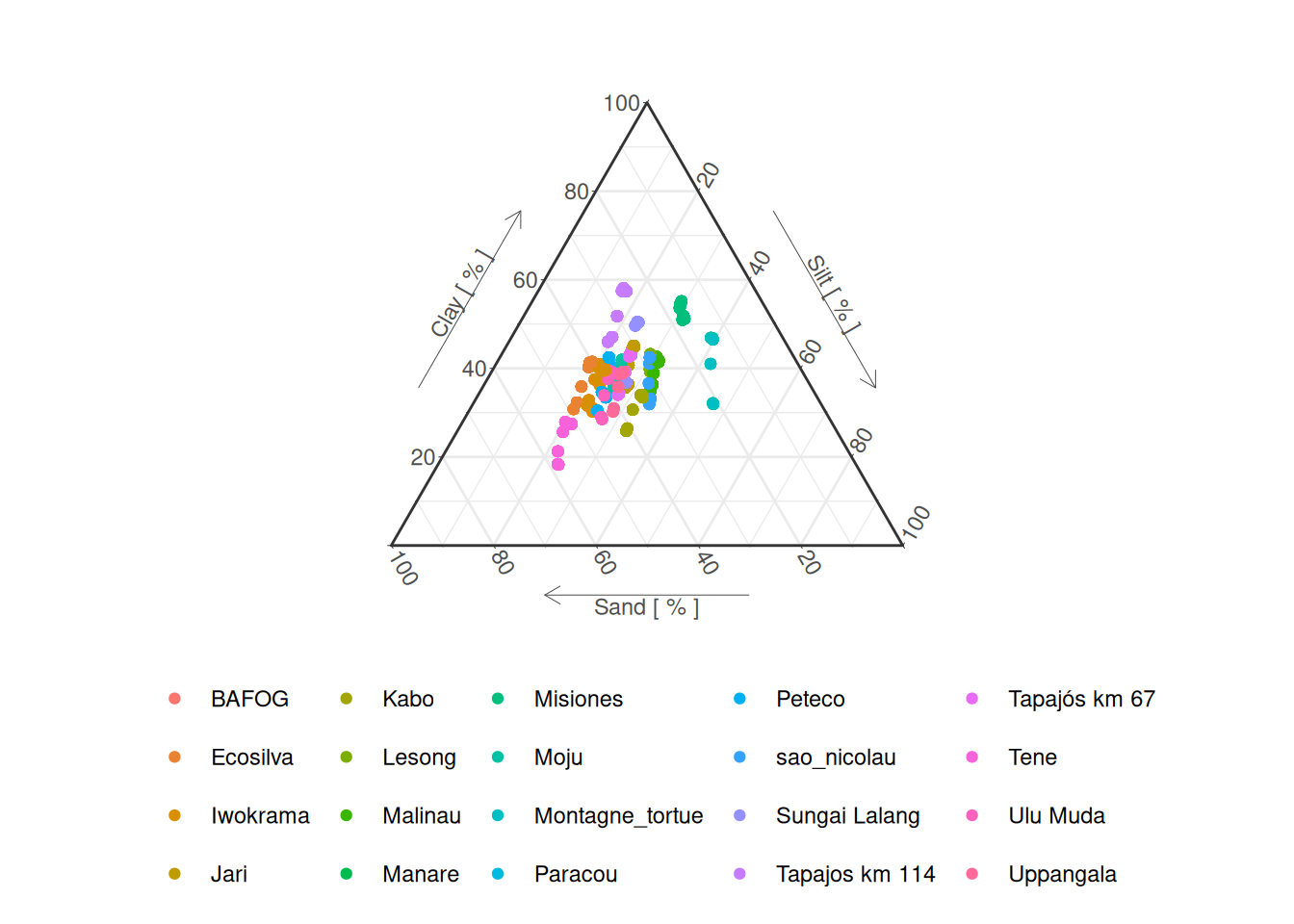
read_tsv ("data/derived_data/soil.tsv" ) %>% ggplot (aes (x = sand, y = clay, z = silt, col = site)) + :: coord_tern (L = "x" , T = "y" , R = "z" ) + geom_point () + theme_bw () + labs (yarrow = "Clay [ % ]" ,zarrow = "Silt [ % ]" ,xarrow = "Sand [ % ]" + :: theme_showarrows () + :: theme_hidetitles () + :: theme_clockwise () + scale_color_discrete ("" ) + theme (legend.position = "bottom" , legend.key.size = unit (0.5 , "line" ))
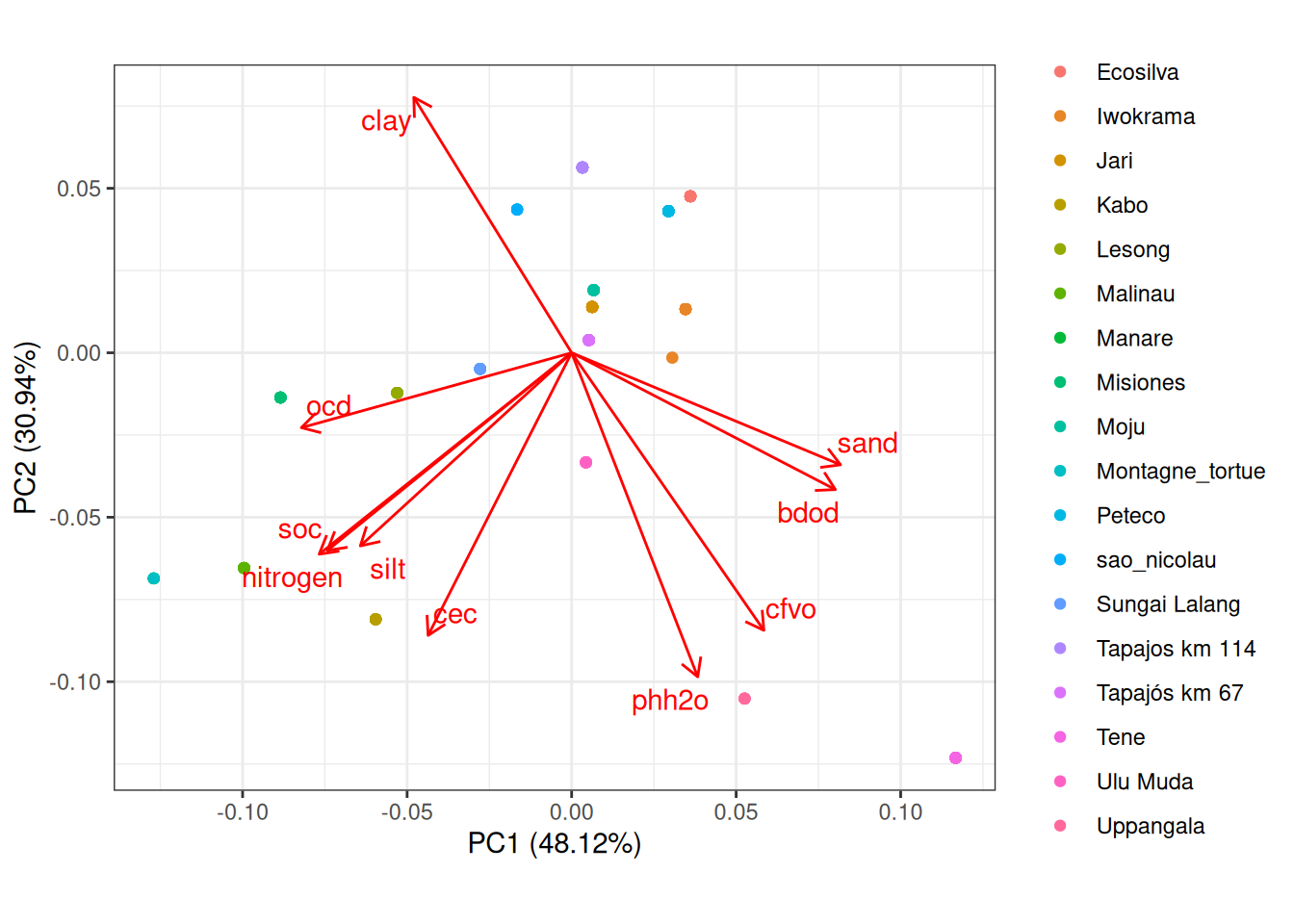
The first ordination axis retained almost half of the variations of the 10 soil variables and opposed sandy poor soils to more fertile silty soils.
Code
library (ggfortify)<- read_tsv ("data/derived_data/soil.tsv" ) %>% select (- X, - Y) %>% group_by (site) %>% filter (depth <= 15 ) %>% summarise_all (mean, na.omit = TRUE ) %>% select (- ocs) %>% na.omit ()<- prcomp (data %>% select (- site, - depth), scale. = TRUE )autoplot (pca,loadings = TRUE , loadings.label = TRUE ,loadings.label.repel = TRUE ,data = data, colour = "site" + theme_bw () + coord_equal () + scale_color_discrete ("" ) + theme (legend.key.size = unit (0.5 , "line" ))
Code
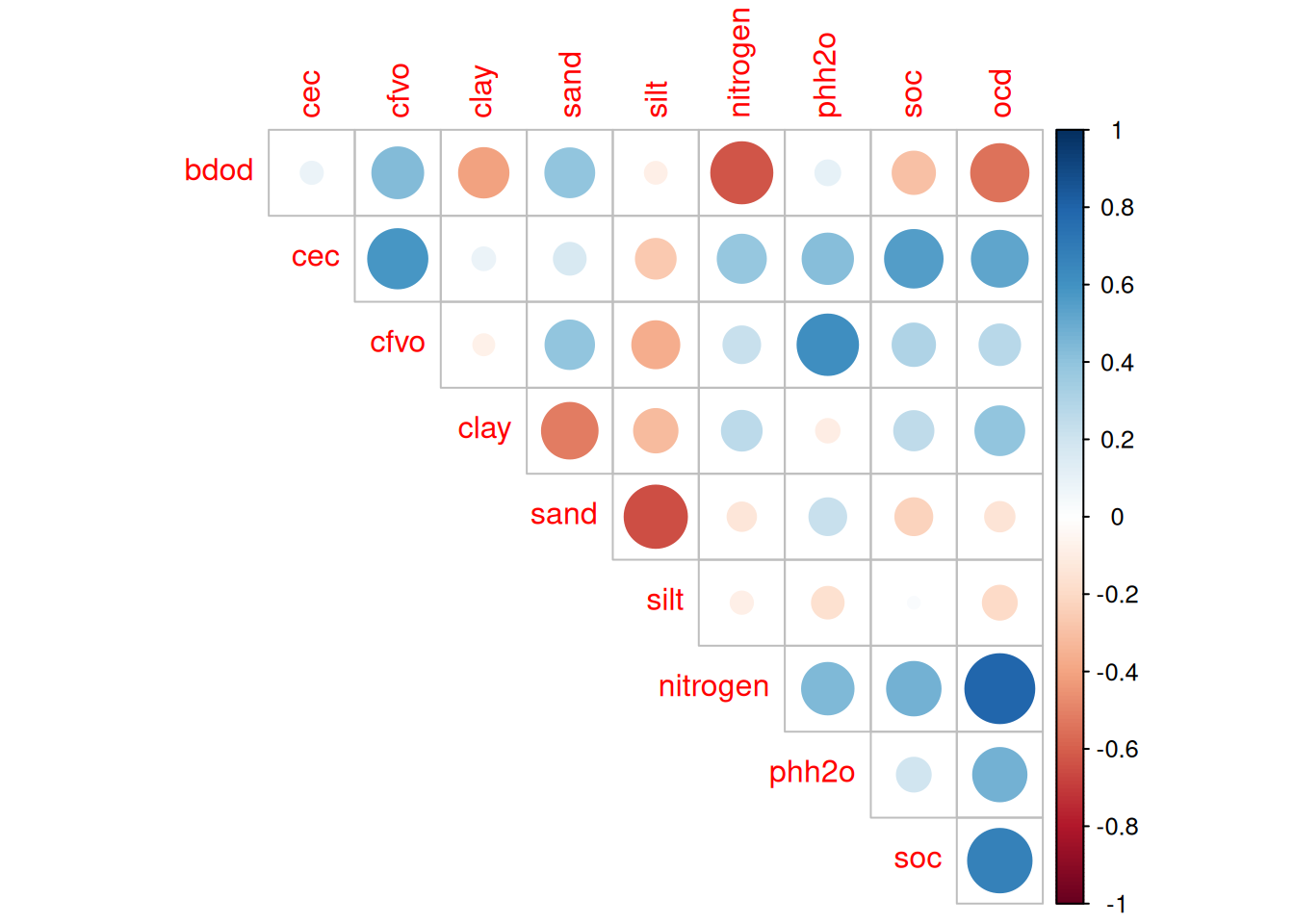
read_tsv ("data/derived_data/soil.tsv" ) %>% select (- X, - Y) %>% group_by (site) %>% filter (depth <= 15 ) %>% summarise_all (mean, na.omit = TRUE ) %>% select (- ocs) %>% na.omit () %>% select (- site, - depth) %>% cor (use = "pairwise.complete.obs" ) %>% :: corrplot (type = "upper" , diag = FALSE )
We will extract all for modelling but group 2 is focused on soil fertility with bulk density (bdod) and soil organic carbon (soc). Note that V. Freycon recommend using cay which is very linked to soil fertility in undistrubed tropical forests as well as to water properties.
Poggio, Laura, Luis M. de Sousa, Niels H. Batjes, Gerard B. M. Heuvelink, Bas Kempen, Eloi Ribeiro, and David Rossiter. 2021.
“SoilGrids 2.0: Producing Soil Information for the Globe with Quantified Spatial Uncertainty.” SOIL 7 (1): 217–40.
https://doi.org/10.5194/soil-7-217-2021 .