Data
Code
plot_info <- read.csv("data/raw_data/bioforest-plot-information.csv") |>
select(site, plot, Year_of_harvest, Treatment)
data <- read.csv("data/derived_data/aggregated_data_v9.csv") |>
mutate(plot_new = Plot) |>
separate(Plot, "Plot", "_") |>
left_join(plot_info, by = join_by(Site == site, Plot == plot)) |>
filter(grepl("cwmdiff", variable)) |>
separate(variable, c("trait", NA, "flux"), sep = "_")
For now, we only calibrate the model with sites that have censuses every one to two years (Paracou, Mbaiki and SUAS).
Code
data <- data |>
filter(Site %in% c("Paracou", "Mbaiki", "SUAS"))
Model
Code
data_rec <- data |>
mutate(trec = Year - Year_of_harvest) |>
filter(Treatment == "Logging" & trec > 3) |>
filter(Site == "Paracou" & trait == "WD" & flux == "all") |>
mutate(plotnum = as.numeric(as.factor(paste(Site, plot_new))))
mdata_cwd <- list(
N = nrow(data_rec),
K = 2,
P = max(data_rec$plotnum),
y = data_rec$value,
plot = data_rec$plotnum,
time = data_rec$trec
)
sample_model("exp_spline", mdata_cwd)
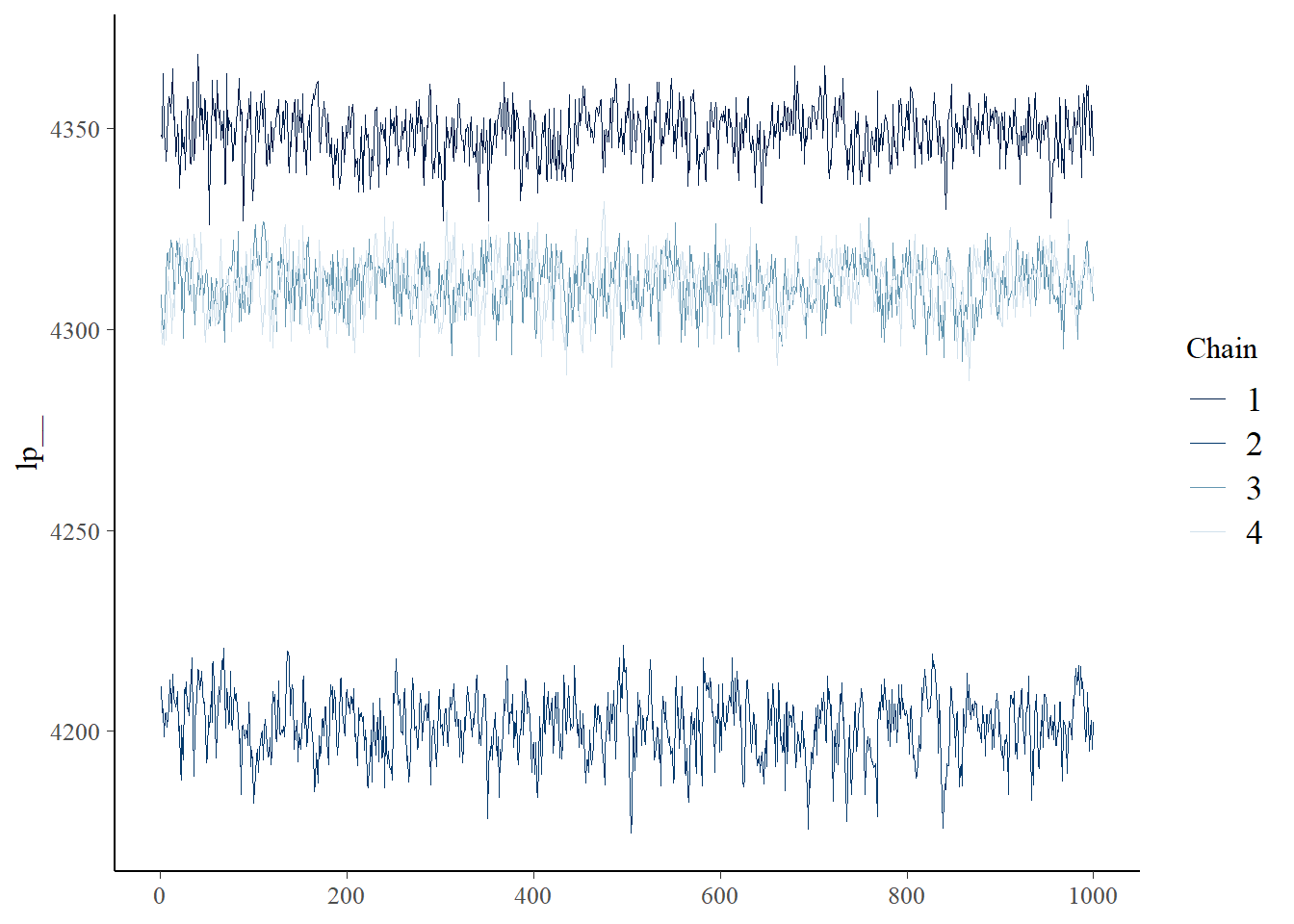
The resulting Stan fit summary is presented below.
Code
fit_flux_cwd <- as_cmdstan_fit(
list.files("chains/exp_spline/", full.names = TRUE)
)
fit_flux_cwd
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
lp__ 4293.25 4311.71 55.60 28.55 4194.96 4354.45 2.41 4 30
sigma 0.00 0.00 0.00 0.00 0.00 0.00 2.32 4 31
dlogtau[1] 2.06 2.15 0.22 0.08 1.67 2.26 1.98 5 33
dlogtau[2] 2.57 2.53 0.56 0.62 1.79 3.40 2.42 4 29
nu[1] 1.25 0.88 0.96 0.54 0.38 2.95 2.10 5 33
nu[2] 3.34 2.96 0.89 0.42 2.52 4.97 2.15 5 33
delta[1,1] -0.01 -0.01 0.00 0.00 -0.01 0.00 1.37 8 28
delta[2,1] -0.01 -0.01 0.00 0.00 -0.01 -0.01 1.05 53 67
delta[3,1] 0.00 0.00 0.00 0.00 0.00 0.00 1.43 8 29
delta[4,1] 0.00 0.00 0.00 0.00 0.00 0.00 1.37 8 29
# showing 10 of 2135 rows (change via 'max_rows' argument or 'cmdstanr_max_rows' option)
Code
mcmc_trace(fit_flux_cwd$draws(variables = c("lp__")))
Code
c("tau", "delta", "nu") |>
fit_flux_cwd$summary()
# A tibble: 76 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 tau[1] 7.99 8.59 1.55e+0 7.23e-1 5.33 9.57e+0 1.98 5.52
2 tau[2] 23.2 21.7 9.56e+0 8.38e+0 11.4 3.84e+1 2.42 4.90
3 delta[1,1] -0.00663 -0.00681 1.23e-3 1.04e-3 -0.00838 -4.21e-3 1.37 8.82
4 delta[2,1] -0.00652 -0.00647 9.16e-4 8.48e-4 -0.00814 -5.12e-3 1.05 53.4
5 delta[3,1] -0.00284 -0.00307 1.31e-3 1.09e-3 -0.00463 -1.95e-4 1.43 8.09
6 delta[4,1] -0.00194 -0.00215 1.25e-3 1.03e-3 -0.00367 5.18e-4 1.37 8.76
7 delta[5,1] -0.00524 -0.00561 1.47e-3 1.11e-3 -0.00707 -2.25e-3 1.50 7.43
8 delta[6,1] -0.00236 -0.00235 8.60e-4 7.86e-4 -0.00376 -9.57e-4 1.03 116.
9 delta[7,1] -0.00571 -0.00589 1.22e-3 1.01e-3 -0.00740 -3.30e-3 1.36 9.09
10 delta[8,1] -0.00446 -0.00462 1.16e-3 9.78e-4 -0.00604 -2.17e-3 1.27 11.1
# ℹ 66 more rows
# ℹ 1 more variable: ess_tail <dbl>
Code
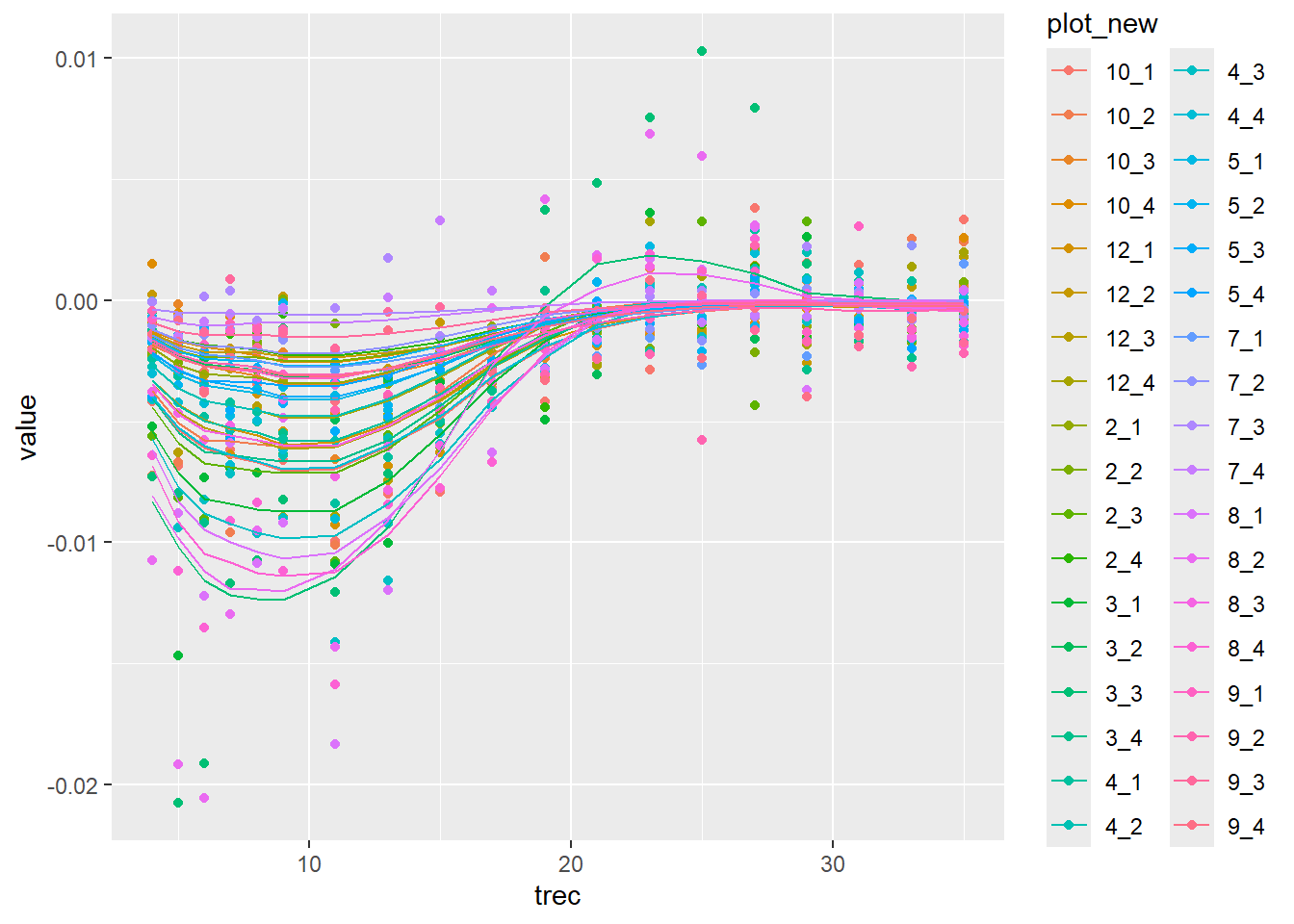
data |>
mutate(trec = Year - Year_of_harvest) |>
filter(Treatment == "Logging" & trec > 3) |>
filter(Site == "Paracou" & trait == "WD" & flux == "all") |>
mutate(med = fit_flux_cwd$summary("mu")$median) |>
mutate(q5 = fit_flux_cwd$summary("mu")$q5) |>
mutate(q95 = fit_flux_cwd$summary("mu")$q95) |>
ggplot(aes(trec, value, col=plot_new)) +
geom_point() +
geom_line(aes(y=med))