data <- read_tsv("../modelling/data/derived_data/data_rec.tsv", col_types = cols()) %>%
select(-y) %>%
filter(variable %in% c("ba", "diversity_q1_gen")) %>%
select(-sitenum, -plotnum) %>%
pivot_wider(names_from = variable, values_from = value) %>%
na.omit() %>%
filter(!(site %in% c("Manare", "Montagne Tortue", "Nelliyampathy",
"Uppangala", "BAFOG", "Sao Nicolau",
"Kabo", "Mil", "Corinto", "Jenaro Herrera",
"Peixoto", "Iwokrama", "Antimary", "Peteco")))
trajectories <- read_tsv("chains/dist-diversity_q1_gen-trajectories.tsv")
parameters <- read_tsv("chains/dist-diversity_q1_gen-parameters.tsv")
area <- data %>%
select(site, plot, area) %>%
unique() %>%
group_by(site) %>%
summarise(area = mean(area))
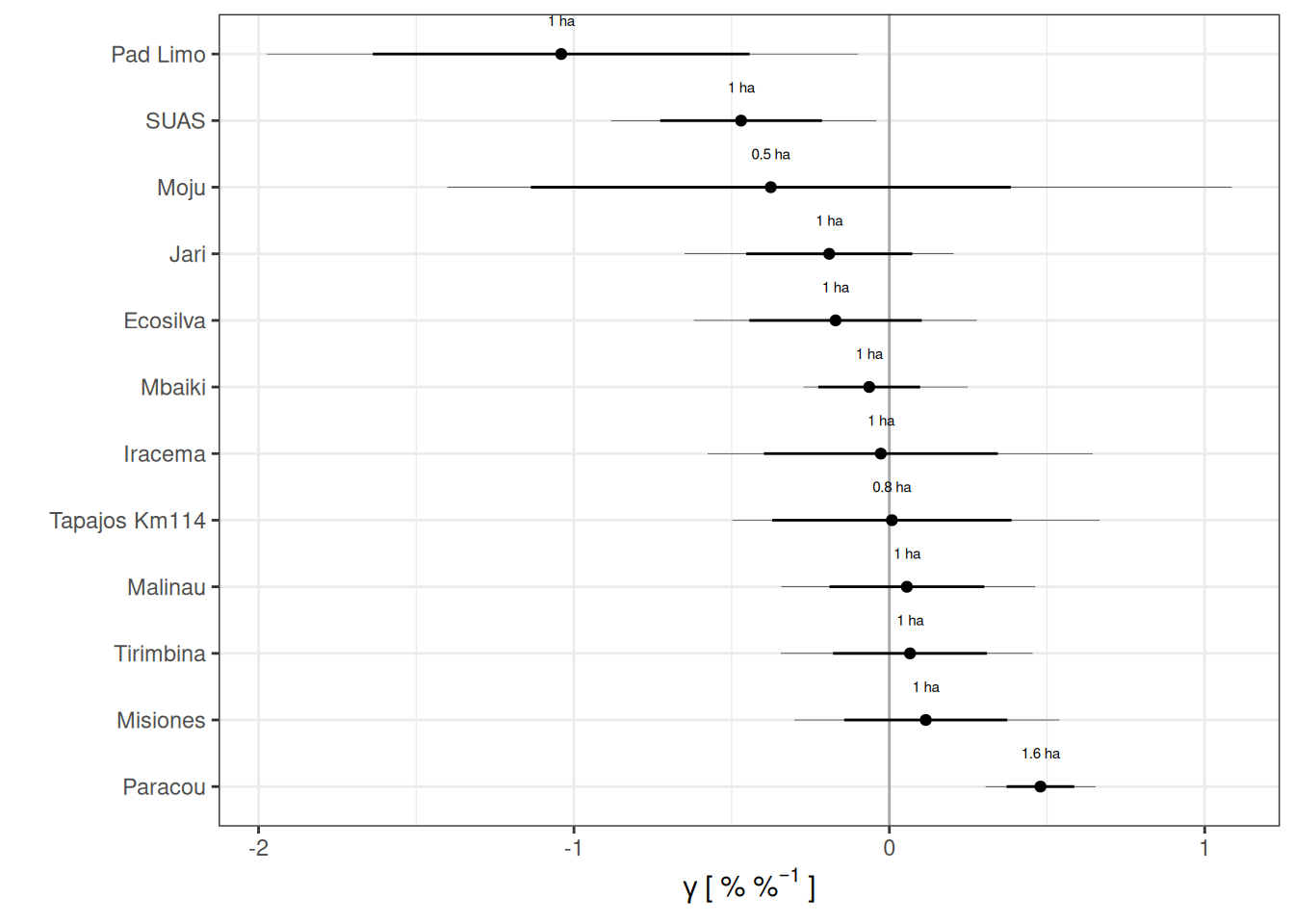
filter(parameters, variable == "gamma") %>%
left_join(area) %>%
ggplot(aes(median, reorder(site, median, decreasing = TRUE))) +
geom_vline(xintercept = 0, col = "darkgrey") +
geom_segment(aes(x = (median - sd),
xend = (median + sd))) +
geom_segment(aes(x = q5, xend = q95), size = .1) +
geom_point() +
geom_text(aes(label = paste(round(area, 1), "ha")), nudge_y = .5, size = 2) +
theme_bw() +
scale_x_continuous(expression(gamma~"["~"%"~"%"^{-1}~"]")) +
ylab("") +
scale_color_viridis_c()