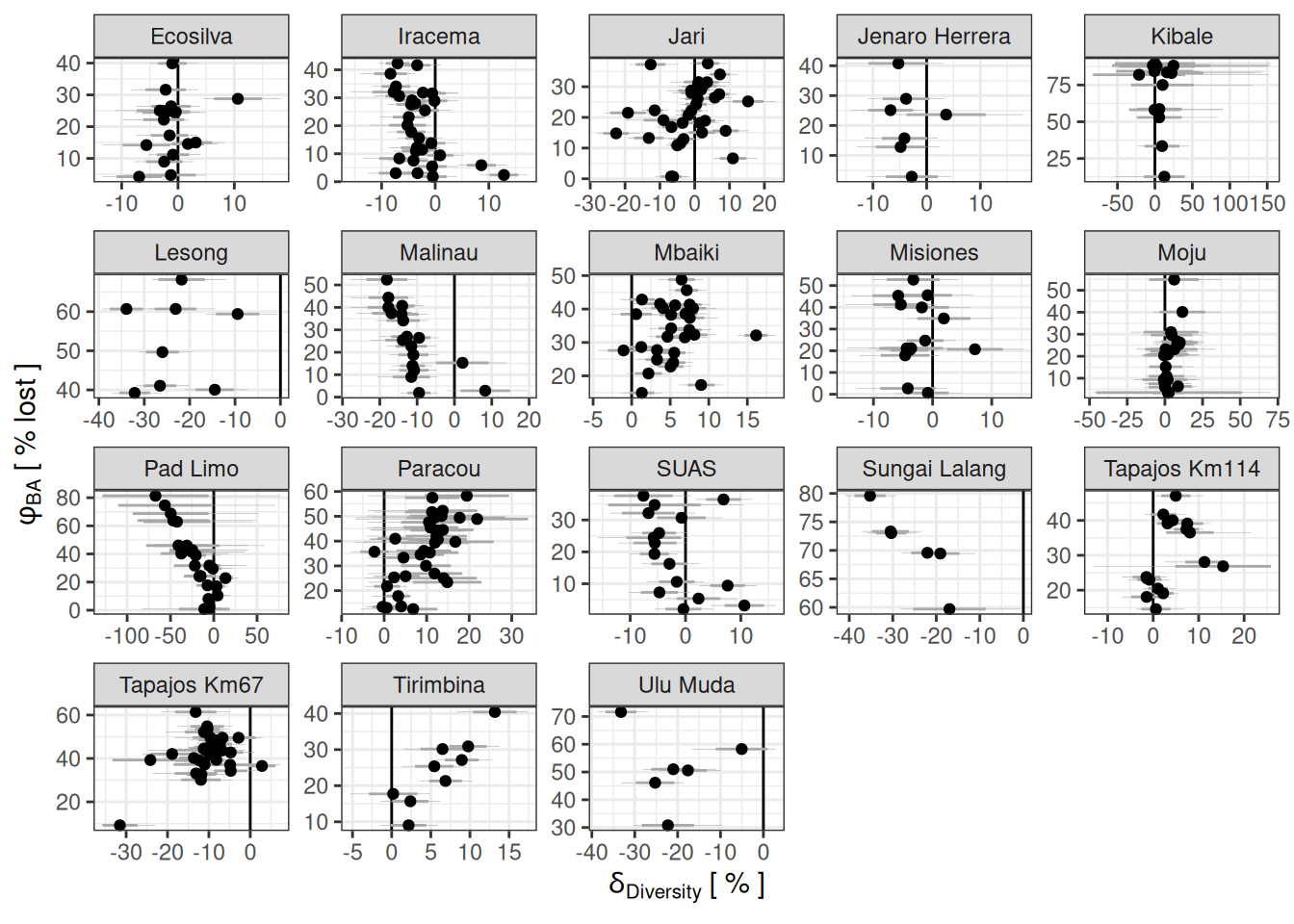
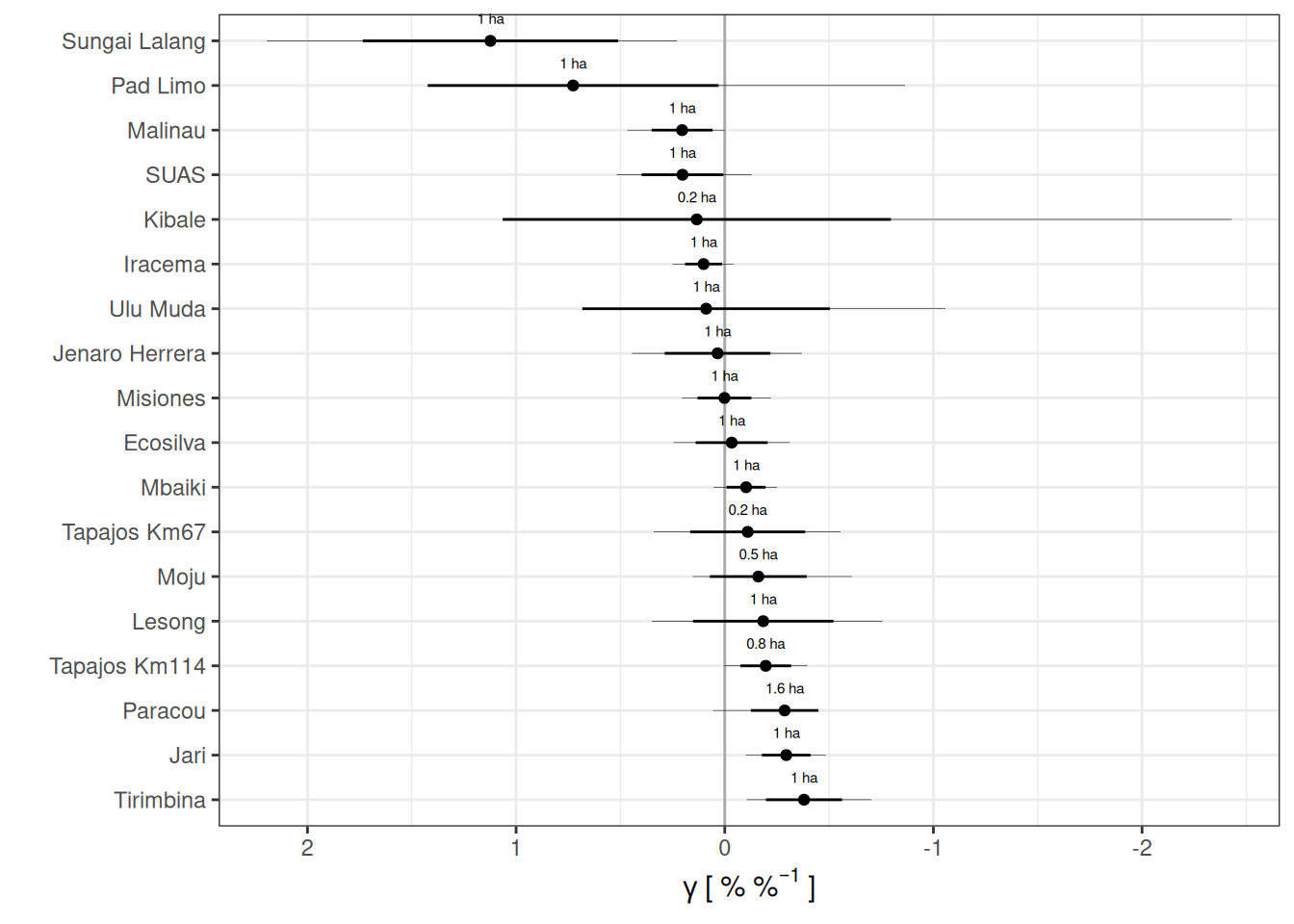
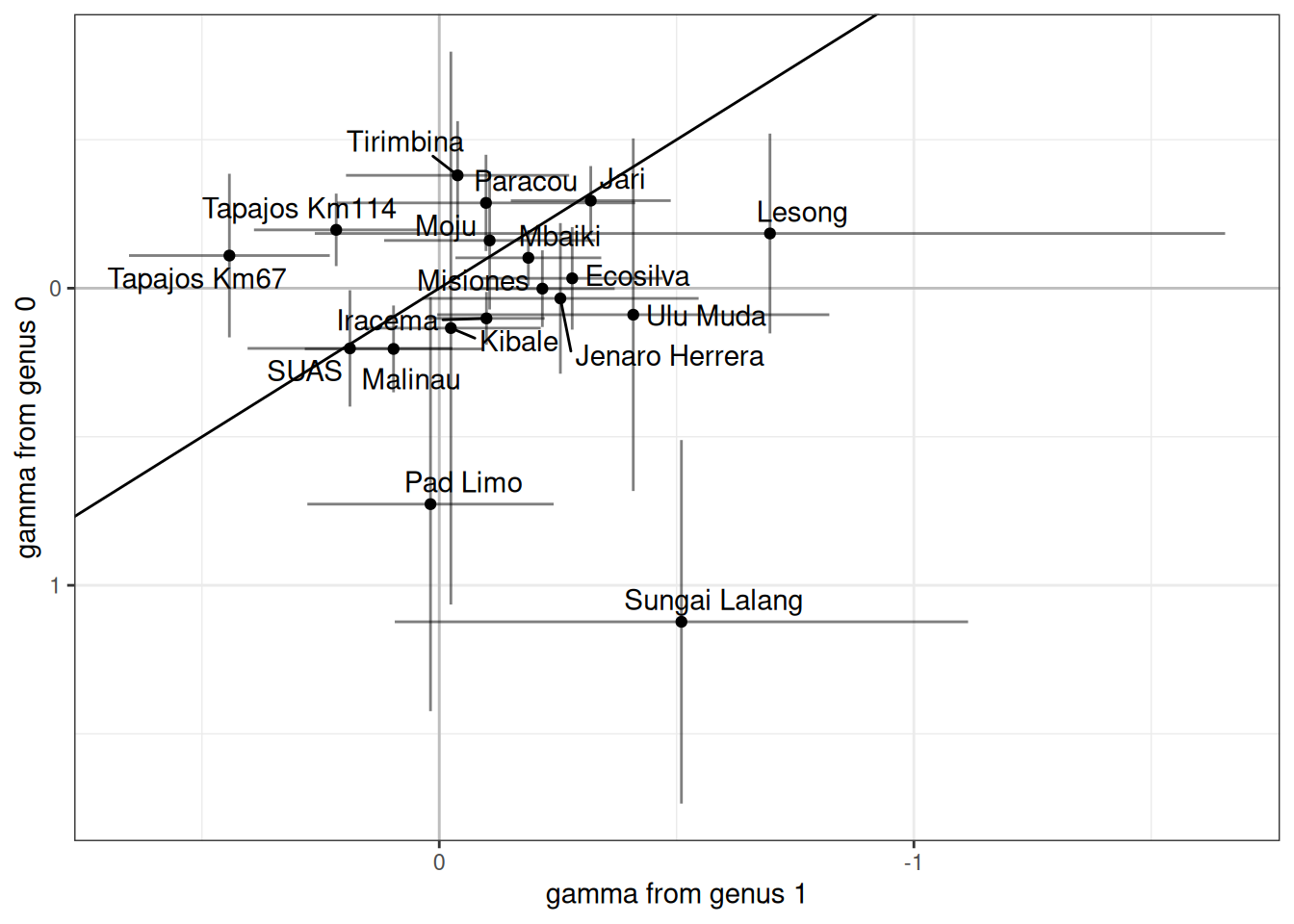
Code
data <- read_tsv("../modelling/data/derived_data/data_rec.tsv", col_types = cols()) %>%
select(-y) %>%
filter(variable %in% c("ba", "diversity_q0_gen")) %>%
select(-sitenum, -plotnum) %>%
pivot_wider(names_from = variable, values_from = value) %>%
na.omit() %>%
filter(!(site %in% c("Manare", "Montagne Tortue", "Nelliyampathy",
"Uppangala", "BAFOG", "Sao Nicolau",
"Kabo", "Mil", "Corinto",
"Peixoto", "Iwokrama", "Antimary", "Peteco"))) %>%
mutate(sitenum = as.numeric(as.factor(site))) %>%
mutate(plotnum = as.numeric(as.factor(paste(site, plot)))) %>%
arrange(sitenum, plotnum, rel_year) %>%
gather(variable, y, -plot, -year, -site, -treatment, -harvest_year,
-longitude, -harvest_year_min, -rel_year, -sitenum, -plotnum)
trajectories <- read_tsv("chains/joint-diversity_q0_gen-trajectories.tsv")
parameters <- read_tsv("chains/joint-diversity_q0_gen-parameters.tsv")
parameters# A tibble: 727 × 14
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ 1.78e+4 1.78e+4 1.11e+2 1.17e+2 1.76e+4 1.80e+4 1.93 5.67
2 gamma -3.40e-2 -3.32e-2 1.73e-1 1.65e-1 -3.12e-1 2.46e-1 1.01 718.
3 gamma 1.03e-1 1.02e-1 8.88e-2 8.58e-2 -4.22e-2 2.51e-1 1.02 234.
4 gamma -2.95e-1 -2.95e-1 1.16e-1 1.13e-1 -4.85e-1 -1.01e-1 1.01 1102.
5 gamma 3.82e-2 3.44e-2 2.53e-1 2.35e-1 -3.70e-1 4.45e-1 1.01 2257.
6 gamma -2.32e-1 1.34e-1 9.30e-1 3.50e-1 -2.43e+0 5.24e-1 1.70 6.26
7 gamma -1.88e-1 -1.84e-1 3.36e-1 3.08e-1 -7.56e-1 3.49e-1 1.02 363.
8 gamma 2.15e-1 2.04e-1 1.46e-1 1.40e-1 -2.97e-4 4.67e-1 1.01 1086.
9 gamma -1.00e-1 -1.02e-1 9.35e-2 9.25e-2 -2.51e-1 5.35e-2 1.00 2626.
10 gamma -2.18e-3 1.42e-3 1.29e-1 1.20e-1 -2.22e-1 2.05e-1 1.00 1735.
# ℹ 717 more rows
# ℹ 5 more variables: ess_tail <dbl>, sitenum <dbl>, site <chr>, plotnum <dbl>,
# plot <chr>